The secrets behind Nanolive’s imaging patented technology
Nanolive’s 3D Cell Explorer’s technology is unique worldwide and is based on a fundamental patent (US 8,937,722 & EU WO 2011/121523).
The combination of holography and rotational scanning makes Nanolive imaging a revolutionary technology.
Holography offers a unique means to measure cells in their native environment: label-free, non-invasive, manipulation-free, and interference-free. Rotational scanning allows 3D reconstructions, noise robustness, and a resolution far beyond the accepted limit for light.
To achieve optimal results for every measurement, the optics of our technology are completely self-adjusting. It guarantees the best alignment conditions – whatever the environment is.

How does the 3D Cell Explorer produce cell images with better than 200 nm lateral resolution?
Classical Microscopy (e.g. fluorescence imaging)
The cell is directly imaged; so what you see on your camera is the image of the cell. Therefore, one absolutely needs to apply Nyquist sampling on the camera. But, this is NOT true for the 3D Cell Explorer. The paradigm shift to make is that there is no direct imaging of the cell on the camera.
Nanolive Imaging
Non-invasive optical nanoscopy can achieve such a lateral resolution by using a quasi-2π-holographic detection scheme and complex deconvolution. The spatial frequencies of the imaged cell do not make any sense to the human eye. But these SCATTERED frequencies are converted into a hologram and synthesize a bandpass which has a resolution DOUBLE the one normally available. Holograms are recorded from different illumination directions on the sample plane and observe sub-wavelength tomographic variations of the specimen. Nanoscale apertures serve to calibrate the tomographic reconstruction and to characterize the imaging system by means of the coherent transfer function. This gives rise to realistic inverse filtering and guarantees true complex field reconstruction.
All the math and proofs are explained in great detail in our peer reviewed Nature Photonics publication here: https://www.nature.com/articles/nphoton.2012.329.
Explaining the Resolution Jump
Rayleigh resolution for incoherent light (e.g. fluo) is 400nm=0.61*520nm/(0.8), for coherent light (e.g. holography) 533nm=0.82*520nm/(0.8) with the diffraction limit (e.g. any imaging by niquist) of 325nm=0.5*520nm/(0.8). In our case for the 3D Cell Explorer configuration, the maximal expected resolution is around 160nm=0.5*520nm/(2*0.8) and we estimate the real resolution (according to Rayleigh) to be about ~190nm. Therefore, taking into account the illuminating wavelength the experimental resolution is 190nm=/lambda/(3.5 NA), indicating a sub-diffraction resolution (d=325 nm at given wavelength and NA) imaging in far-field (non-fluorescent) transmission microscopy.
In conclusion, the 2 terminologies of (i) optical resolution (the real one) and (ii) sampling resolution (the one on the screen) are separated for 3D holotomographic microscopy. For the sake of computational cost-reduction, Nanolive chose to have the sampling optimal at the optical resolution, hence about 180 nm. Again, we can do this, thanks to our technology WITHOUT loosing any real resolution, but of course a user could over-sample the images and have double/triple/etc sampling on the screen with the same optical resolution (e.g. simple zero-padding in fft space to have a smoother image).
The images
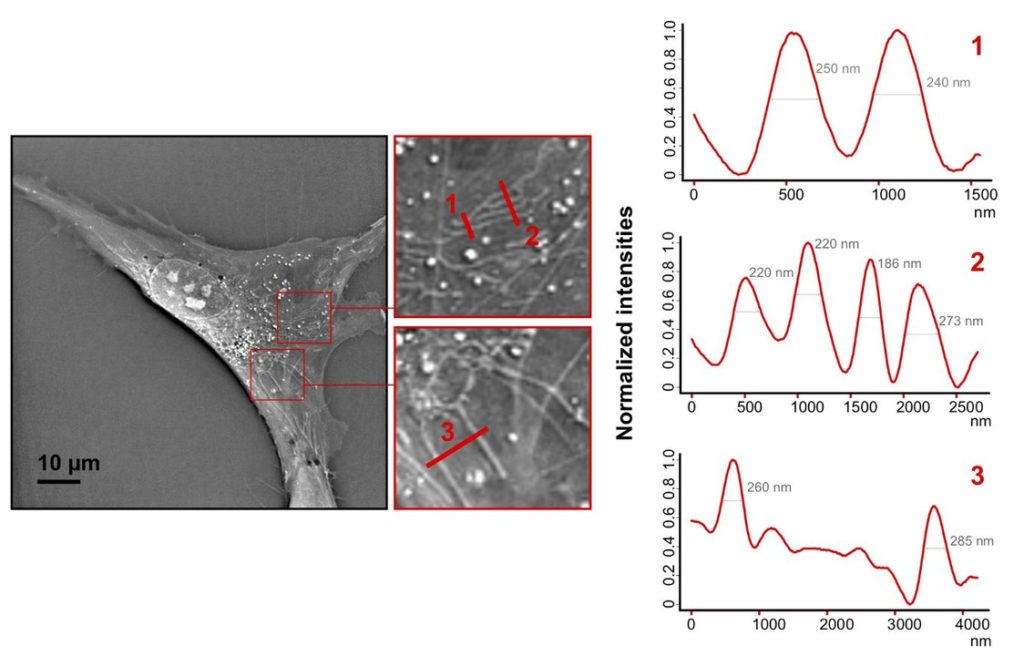
We have chosen Mitochondria as a great candidate to demonstrate resolution. Below is an image calculating the separation between Mitochondria using standard signal profiling. In the example we resolve mitochondria tubules as thin as 186 nm (using full width at half maximum standard measurement of object thickness).
What fixation does to your cells’ morphology
In life sciences, fixation refers to the process of preservation of histological, cytological and microbiological specimens such as cells and tissues at the status they were originally taken. A protocol of fixation usually precedes immunostaining, which is essential to immobilize biological structures and make them accessible to antibodies.
There are different mechanisms of fixation depending on the reagent used. As already shown in a previous publication from Sandoz et al (Fig.2A), dehydrating fixation methods severely affected organelles structure preservation and thereby their RI. A widely used alternative is paraformaldehyde (PFA), which acts creating covalent chemical cross-links between proteins. Even if PFA rapidly penetrates the sample, the process can take up to a few weeks in case of human biopsies for example.
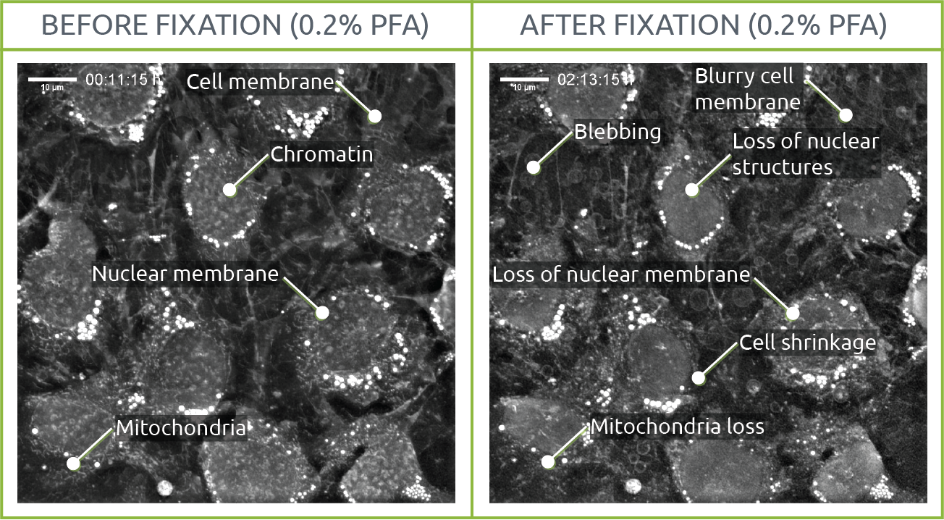
Since fixation aims to provide a snapshot of a living situation, we visualized the process of fixing living samples with PFA using Nanolive’s 3D Cell Explorer. We conclude that cell structures are affected by the fixation process.
Two samples of living mouse pre-adipocytes were imaged for 30 min before initiating the fixation process. Images were taken every 15 seconds both before and after fixation. On the 0.2% PFA condition, PFA was directly diluted in medium with a final concentration of 0.2% PFA at 30min. On the 2% PFA condition, the medium was completely removed and replaced by 2% PFA in PBS after 30min. The process of fixation was recorded for up to 1 hour.
0.2% PFA
The effects of fixation could be observed early on at the level of cellular structures. In this sense, while chromatin, nuclear membrane and mitochondria were distinguishable before fixation, their shape and density were affected after PFA was added to the medium. This same phenomenon was observed on the cellular membrane, with cells shrinking. Several blebs appeared throughout the sample.
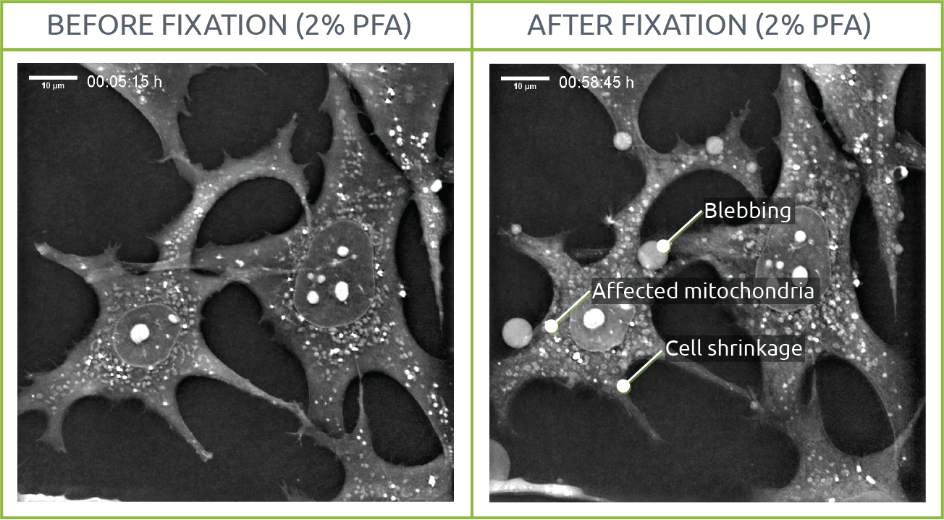
2% PFA
Morphological changes such as variations in mitochondrial shape and density or cell shrinkage were visualized. In a similar way to the cells observed on the 0.2% PFA, membrane degradation led to blebbing.
Interestingly, mitochondria shape evolved into almost perfect rounded shapes after fixation. A phenomenon of “granularity” was also observed in those organelles: while mitochondrial content lost density, their membrane looked “brighter” than before fixation probably due to fixation-derived changes in membrane nature and integrity.
Highlights
All in all, these observations strongly question the use of fixed samples in research involving membrane dependent systems.
A concentration as low as 0.2% PFA already induces very strong cellular changes at the molecular level, which are translated in an observation that does not meet the characteristics of living cells. These modifications are much more accentuated amongst membranes (mitochondrial, nuclear and plasma).
These conclusions correlate with previous research on the subject[1], [2]. PFA fixation has been shown to induce strong perturbations of cellular nanostructures[2], and inefficiently fix glycosylphosphatidylinositol-anchored, phospholipid-anchored or cholesterol-anchored membrane proteins, leading to observation of aberrant clustering of proteins observed by subsequent immune-staining[1].
[1] K. A. K. Tanaka et al., “Membrane molecules mobile even after chemical fixation,” Nature Methods, vol. 7, no. 11. pp. 865–866, Nov-2010.
[2] Y. Li et al., “The effects of chemical fixation on the cellular nanostructure,” Exp. Cell Res., vol. 358, no. 2, pp. 253–259, Sep. 2017.
The secrets behind Nanolive’s imaging patented technology
Classical Microscopy (e.g. fluorescence imaging)
The cell is directly imaged; so what you see on your camera is the image of the cell. Therefore, one absolutely needs to apply Nyquist sampling on the camera. But, this is NOT true for the 3D Cell Explorer. The paradigm shift to make is that there is no direct imaging of the cell on the camera.
Nanolive Imaging
Non-invasive optical nanoscopy can achieve such a lateral resolution by using a quasi-2π-holographic detection scheme and complex deconvolution. The spatial frequencies of the imaged cell do not make any sense to the human eye. But these SCATTERED frequencies are converted into a hologram and synthesize a bandpass which has a resolution DOUBLE the one normally available. Holograms are recorded from different illumination directions on the sample plane and observe sub-wavelength tomographic variations of the specimen. Nanoscale apertures serve to calibrate the tomographic reconstruction and to characterize the imaging system by means of the coherent transfer function. This gives rise to realistic inverse filtering and guarantees true complex field reconstruction.
All the math and proofs are explained in great detail in our peer reviewed Nature Photonics publication here: https://www.nature.com/articles/nphoton.2012.329.
Explaining the Resolution Jump
Rayleigh resolution for incoherent light (e.g. fluo) is 400nm=0.61*520nm/(0.8), for coherent light (e.g. holography) 533nm=0.82*520nm/(0.8) with the diffraction limit (e.g. any imaging by niquist) of 325nm=0.5*520nm/(0.8). In our case for the 3D Cell Explorer configuration, the maximal expected resolution is around 160nm=0.5*520nm/(2*0.8) and we estimate the real resolution (according to Rayleigh) to be about ~190nm. Therefore, taking into account the illuminating wavelength the experimental resolution is 190nm=/lambda/(3.5 NA), indicating a sub-diffraction resolution (d=325 nm at given wavelength and NA) imaging in far-field (non-fluorescent) transmission microscopy.
In conclusion, the 2 terminologies of (i) optical resolution (the real one) and (ii) sampling resolution (the one on the screen) are separated for 3D holotomographic microscopy. For the sake of computational cost-reduction, Nanolive chose to have the sampling optimal at the optical resolution, hence about 180 nm. Again, we can do this, thanks to our technology WITHOUT loosing any real resolution, but of course a user could over-sample the images and have double/triple/etc sampling on the screen with the same optical resolution (e.g. simple zero-padding in fft space to have a smoother image).
| Properties |  | Standard Fluorescence | Super resolution Fluorescence | Phase imaging | Electrical impedance | SEM, AFM |
| Preparation | ||||||
| Time | ||||||
| Simplicity |  |  |  |  |  |  |
| Cost | ||||||
| Cell condition | ||||||
| Observation | ||||||
| Temporal sampling | ||||||
| Live Cell Imaging | ||||||
| Training-free | ||||||
| Cellular health | ||||||
| Result | ||||||
| Lateral Resolution | Below 200nm | Down to 250nm | Down to 50nm | 400 – 1000nm | No Image | 5-10nm |
| Image Dimensionality | ||||||
| Quantitative | ||||||
| Comprehensive | ||||||
| Costs | ||||||
| Price (euro) |  |
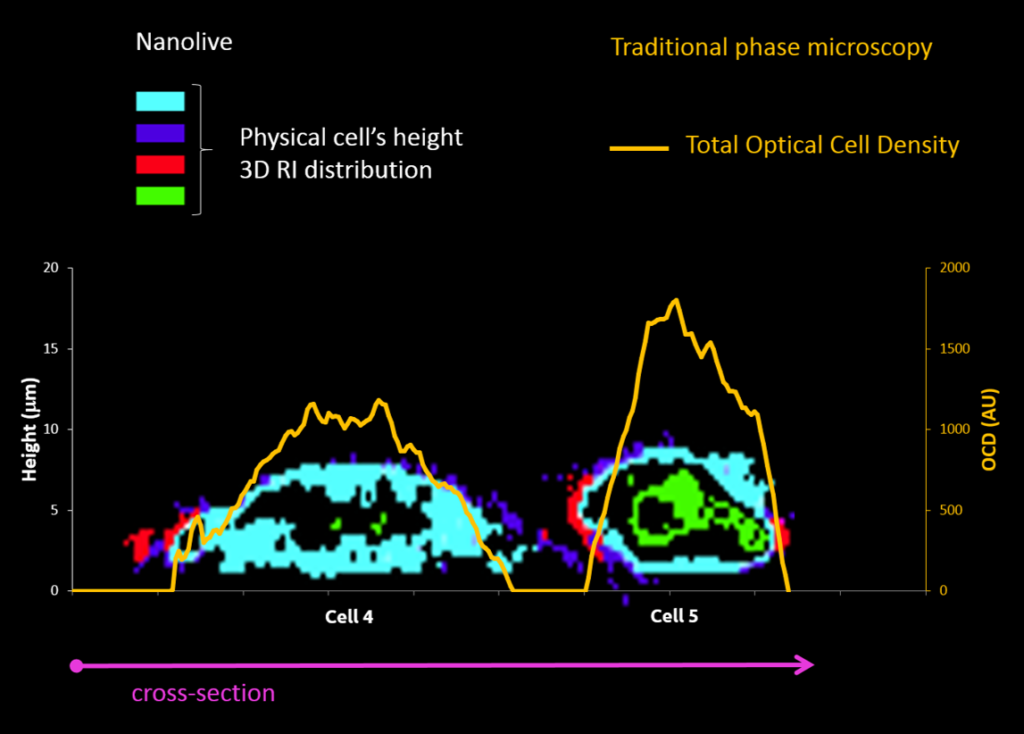
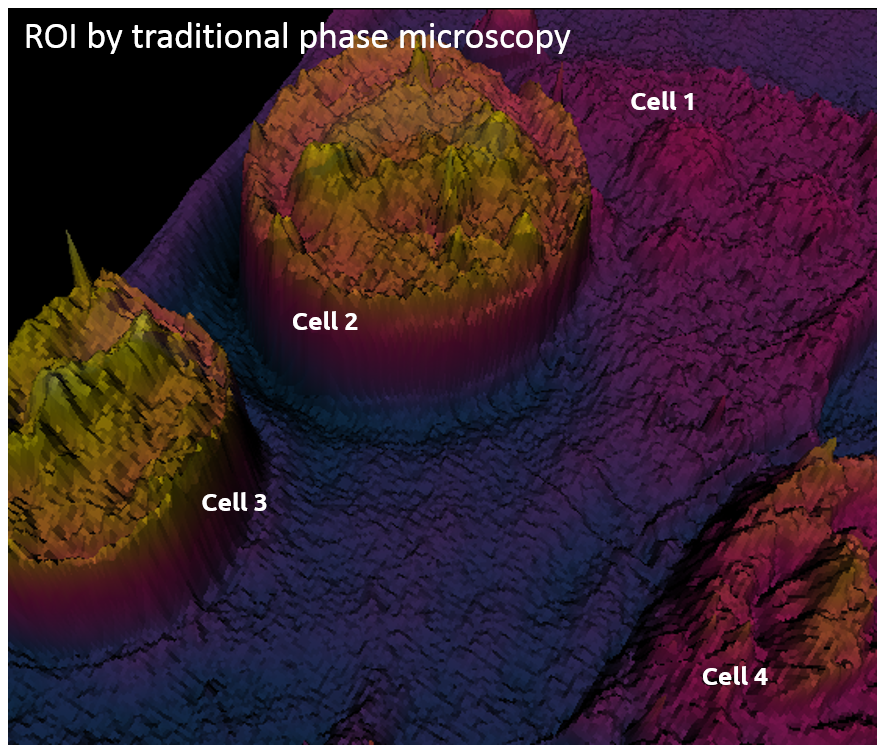
Traditional phase microscopy |
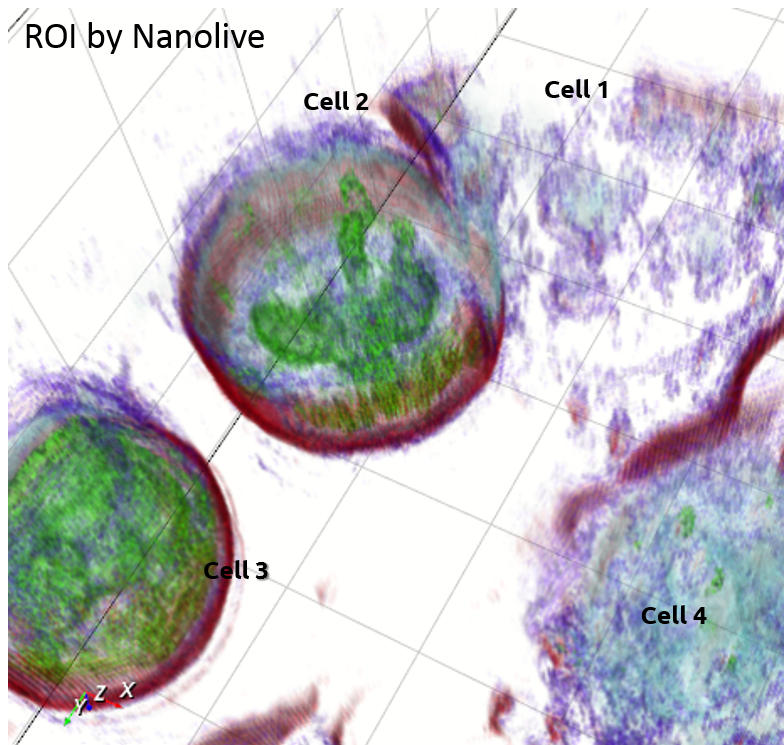
Nanolive |
 |
 |
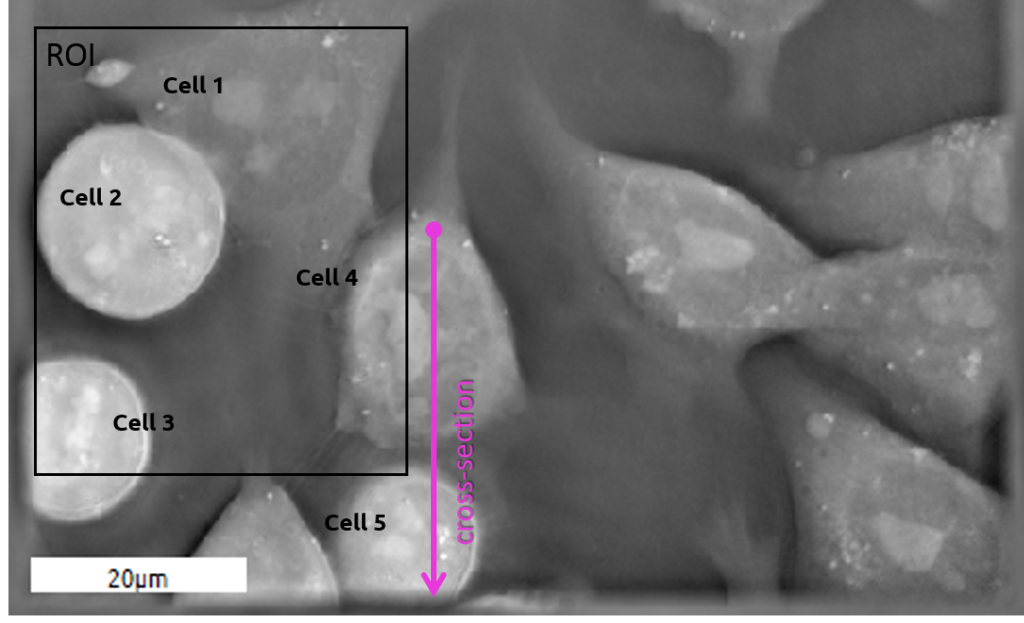
Example of Embryonic Stem Cells (ESC) during mitosis
 |
 |
 |
| 2D RI map of mouse Embryonic Stem Cell Cells (mESC) |
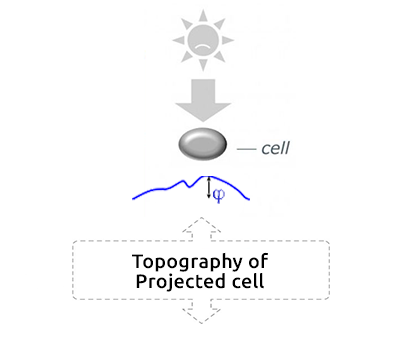
Cell projection (outside cell topography)
Total Optical Cell Density (2.5D OCD)
Resolution: 400 – 1000 nm
High noise level
|
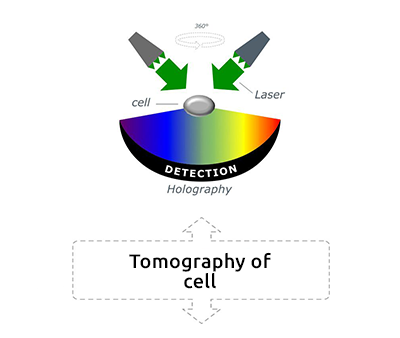
Tomographic Imaging
Physical cell’s height (3D RI distribution) 200 nm resolution Low noise ratio |
Comparison between ESCs (4 & 5) as observed with the two technologies